Pinaki Bose and his team employ spatial transcriptomics to analyze heterogeneity in head and neck cancers, such as OSCC.
Ali Alsaady
The tumor microenvironment (TME) is an intricate ecosystem composed of malignant cells, fibroblasts, immune cells, and endothelial cells.1 Adding to this complexity, the cancerous cells present in the TME are not all identical, where phenotypically distinct malignant cell subpopulations arise through genetic and epigenetic modifications. This intratumoral heterogeneity has a huge effect on cancer progression, metastasis, and a patient’s response to therapy, highlighting the need for further investigation into this aspect of cancer biology.2
Scientists commonly use single-cell technologies, such as RNA sequencing or flow cytometry, to study intratumoral heterogeneity.3 However, these techniques require tissue disassociation and, consequently, do not provide spatial information about the positioning of the cellular subpopulations within the tumor. In a recently published Nature Communications paper, researchers overcame this limitation by employing spatial biology techniques to investigate transcriptional heterogeneity in the TME.4
Pinaki Bose, a cancer researcher at the University of Calgary and corresponding author of this paper, studies head and neck cancers including the most common of these called oral squamous cell carcinoma (OSCC). “Almost 300,000 individuals are afflicted by [OSCC] every year and up to 50 percent die of the disease,” said Bose. “It is also one of the most common cancers in South Asia. It is the number one cancer in India. So, it is a huge disease burden.”
Almost 300,000 individuals are afflicted by [OSCC] every year and up to 50 percent die of the disease. … So, it is a huge disease burden.
Pinaki Bose, University of Calgary
To assess tumoral heterogeneity, Bose and his team prepared slides from 12 OSCC tumor samples resected from 10 patients. They next performed spatial transcriptomics using a technique that sequenced over 24,000 individual spots across each slide. After grouping together malignant spots with similar gene expression profiles, they found that the spots sorted into three major clusters, each with their own unique geographical location in the TME. The researchers identified the upregulated genes in each cluster and compared that data to existing information on OSCC tumor architecture. They concluded that the clusters corresponded to previously discovered regions of OSCC tumors: the tumor core (TC), transitory region, and leading edge (LE).5 Although scientists have studied these regions, they had never been comprehensively characterized.
The researchers then evaluated the similarities between the TC and LE regions from the 12 patient tissue sections in terms of their transcriptional profiles. They found that gene expression was highly correlated in the TC region and the LE region across patients, which suggests that TC and LE transcriptional profiles are conserved among OSCC tumors.
Because genes associated with invasion and metastasis were upregulated in the LE region, Bose and his team wondered if there was a connection between this transcriptional profile and patient prognosis. To do this, they utilized information from The Cancer Genome Atlas Program, a publicly funded project that molecularly profiled various cancers, and analyzed transcriptomic and survival data derived from the same patients. After including data from 20 different cancer types in their analyses, the researchers found that most patients with tumors highly expressing the LE signature had reduced survival outcomes, indicating that this profile could be predictive of poor prognosis in cancers beyond OSCC.
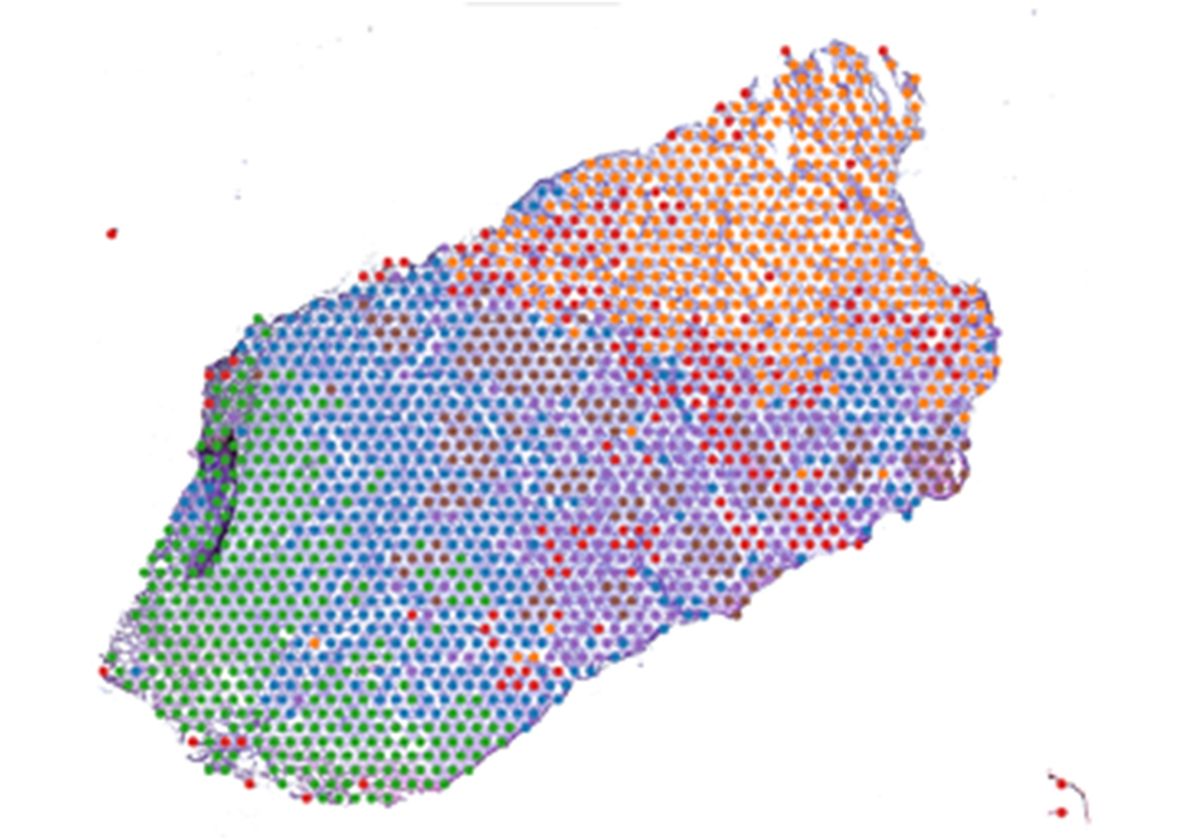
Pinaki Bose and colleagues examined gene expression at thousands of spots localized across hematoxylin and eosin-stained OSCC tissue sections.
Mehul Kumar
“This was a really unique way to integrate a lot of different work that’s been done in cancer biology,” said Kristin Beaumont, a genomic scientist at Icahn School of Medicine at Mount Sinai, who was not involved in the study. “I think that people have certainly studied the individual cellular function of different compartments of the tumor and people have studied the architecture of tumors. But this study, in my mind, helped contribute to the overlap and the integration of those particular directions of investigation.”
Bose credits his talented team for their remarkable findings, including co-first authors Rohit Arora and Christian Cao, who were undergraduate students at the time. “A lot of the analysis was done very independently by them, and it was amazing what we were able to discover from this study,” Bose said. Arora is now a doctoral student at Harvard University, while Cao is a medical student at the University of Toronto.
Bose plans to perform spatial transcriptomic profiling of more OSCC tumors and other cancer types to see if he can identify the LE signature. Additionally, he intends to further analyze available datasets. “There is so much spatial transcriptomics data now to analyze, including ours, that is now online. So, I want to integrate all of that data and do a pan-cancer analysis and understand more about the mechanisms that lead to invasion, metastasis, [and] immune evasion,” explained Bose. “These are big questions and will take a long time to answer, but … we hope to contribute to that feat.”
References
- Tammela T, Sage J. Investigating tumor heterogeneity in mouse models. Annu Rev Cancer Biol. 2020;4(1):99-119.
- Lawson DA, et al. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. 2018;20(12):1349-1360.
- de Vries NL, et al. Unraveling the complexity of the cancer microenvironment with multidimensional genomic and cytometric technologies. Front Oncol. 2020;10.
- Arora R, et al. Spatial transcriptomics reveals distinct and conserved tumor core and edge architectures that predict survival and targeted therapy response. Nat Commun. 2023;14(1):5029.
- Sharma M, et al. Molecular changes in invasive front of oral cancer. J Oral Maxillofac Pathol JOMFP. 2013;17(2):240-247.